Effect of 23‑hydroxybetulinic acid on lung adenocarcinoma and its mechanism of action
- Authors:
- Published online on: March 28, 2024 https://doi.org/10.3892/etm.2024.12527
- Article Number: 239
-
Copyright: © Tan et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
Lung cancer is one of the malignant tumors with the highest incidence and mortality rate worldwide (1). Globally, the number of deaths caused by lung cancer accounts for ~30% of all cancer-associated deaths. In China, lung cancer has the highest incidence and mortality rate among all tumors. The primary treatment for lung cancer is surgical intervention, but with the advancement of medical technology, various treatment methods are emerging. These methods range from conventional chemotherapy to molecular targeted therapies and immunotherapy, such as monoclonal antibodies against EGFR, anaplastic lymphoma kinase (ALK), VEGFR and programmed cell death protein 1/programmed cell death ligand 1 (2,3). Consequently, the treatment of lung cancer is gradually transitioning into the era of precision medicine.
The gene mutation rate in cancerous lung tissues is higher than that of adjacent tissues, indicating that gene mutations could lead to the occurrence of lung cancer and increase its incidence (4,5). Early detection of high calmodulin 1 expression and EGFR mutations can be used to differentiate the diagnosis of patients with colorectal cancer, which is conducive to further prolonging the survival period and improving the quality of life of patients (6). Identifying the p53 Arg72Pro mutation in patients with primary hepatocellular carcinoma is important for the early diagnosis of cancer in patients with chronic hepatitis B-associated cirrhosis (7). Thus, identifying new gene mutation sites and searching for novel targeted therapeutic targets are necessary in the era of precision medicine. The discovery and determination of the role of specific genes in the development of lung cancer is critical in obtaining ideal, effective and precise therapeutic targets (8). Therefore, at present, gene research on lung adenocarcinoma represents a significant focus. EGFR, KRAS, ALK, c-MET, and other genes have been implicated in the progression of lung adenocarcinoma (9), guiding its clinical applications. However, several patients develop drug resistance and gene mutations following targeted therapy for lung adenocarcinoma (10). 23-Hydroxybetulinic acid (23-HBA) has been shown to promote the apoptosis of tumor cells by inhibiting the proliferation of vascular endothelial cells (11). Hence, genetic changes related to lung adenocarcinoma and the inhibitory effects of novel compounds must be further explored (12).
23-HBA is a triterpenoid compound isolated from Pulanemone (13), a traditional Chinese medicine used for heat clearance and detoxification (14). 23-HBA has been demonstrated to reduce the mitochondrial membrane potential and induce apoptosis in melanoma cells (B16), and activate the p38 and JNK MAPK pathway-associated kinases in colon cancer cells (LoVo) (15). The loss of mitochondrial membrane potential in LoVo cells can trigger apoptosis through the mitochondrial pathway (16). Moreover, angiogenesis can be hindered by blocking human microvascular endothelial cells (HMECs) in the S phase (17), restraining HMEC growth and inducing apoptosis. 23-HBA exhibits a synergistic effect with antitumor drugs such as 5-fluorouracil (18). It can also inhibit the activity of the ATP binding cassette subfamily B member 1 ATPase (19), leading to the accumulation of drugs (20) within cells and effectively reversing P-glycoprotein-mediated multidrug resistance. In addition, 23-HBA can induce beclin-1-dependent autophagy and apoptosis (21) in HL-60 cells. 23-HBA has been structurally modified to enhance its antitumor efficacy and bioavailability, resulting in >300 derivatives. Some of these derivatives have shown improved antitumor activity (22). 23-HBA can inhibit the activation of NF-κB p65 and the MAPK pathway, and reduce the secretion of inflammatory factors in mice with DSS-induced acute ulcerative colitis (23). Furthermore, 23-HBA has been shown to reduce tumorigenesis, metastasis and immunosuppression in mouse models of hepatocellular carcinoma by disrupting MAPK signaling pathways (20).
In the present study, the aim of this study is to investigate the underlying mechanisms of action of Pulsatilla compounds against lung adenocarcinoma. The Pulsatilla compounds were combined with lung adenocarcinoma, and databases, such as SwissTargetPrediction, were utilized to demonstrate a robust association between the target genes of 23-HBA and the pathogenesis of lung adenocarcinoma (24). MTT and flow cytometry assays were conducted to validate the effect of 23-HBA on lung adenocarcinoma cells (20). Target genes of 23-HBA in lung adenocarcinoma were identified by bioinformatics analysis and characterized to test their functions. The current study introduces novel perspectives for the treatment of patients with lung adenocarcinoma (25).
Materials and methods
Cell line culture
The human lung adenocarcinoma cell lines A549, H1975 and H1299, as well as the human bronchial epithelial cell line BEAS-2B, were used in the present study. These cell lines were provided by The Cell of Bank of Type Culture Collection of the Chinese Academy of Sciences. The cell lines used in the presented study were tested for mycoplasma contamination, ensuring that the experiments were conducted with uncontaminated cell cultures. Cells were cultured in DMEM High Glucose (Hyclone; Cytiva) with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.). The cells were cultivated in a cell incubator at 37˚C in a humidified atmosphere with 5% CO2.
Antibodies and drugs
Rabbit anti-human GAPDH antibody (1:6,000 dilution; cat. no. AP0063) was purchased from Bioworld Technology, Inc. Peroxisome proliferator-activated receptor (PPAR)-γ rabbit polyclonal antibody (1:1,000 dilution; cat. no. 16643-1-AP) was purchased from Proteintech Group, Inc. HRP-conjugated goat anti-rabbit IgG (1:6,000 dilution; cat. no. BS13278) was purchased from Bioworld Technology, Inc. and was used as a secondary antibody (26).
23-HBA (Shanghai Aladdin Biochemical Technology Co., Ltd.) was used in the present study. It is known by the chemical names is ‘(3β)-3,23-Dihydroxylup-20(29)-en-28-oic acid’ or ‘(1R,3aS,5aR,5bR,7aR,9S,11aR,11bR,13aS,13bR)-9-hydroxy-8-(hydroxymethyl)-5a,5b,8,11a-tetramethyl-1-prop-1-en-2-yl-1,2,3,4,5,6,7,7a,9,10,11,11b,12,13,13a,13b-hexadecahydrocyclopenta[a]chrysene-3a-carboxylic acid’. The molecular weight of 23-HBA is 472.7 g/mol, while it has a melting point of 305˚C. 23-HBA is soluble in dimethyl sulfoxide (DMSO), a commonly used solvent in the laboratory, at a concentration of 2 mg/ml. 23-HBA is sensitive to light, humidity and heat, and its sensitivity is an important point to consider during its handling and storage (27).
Data sources and processing
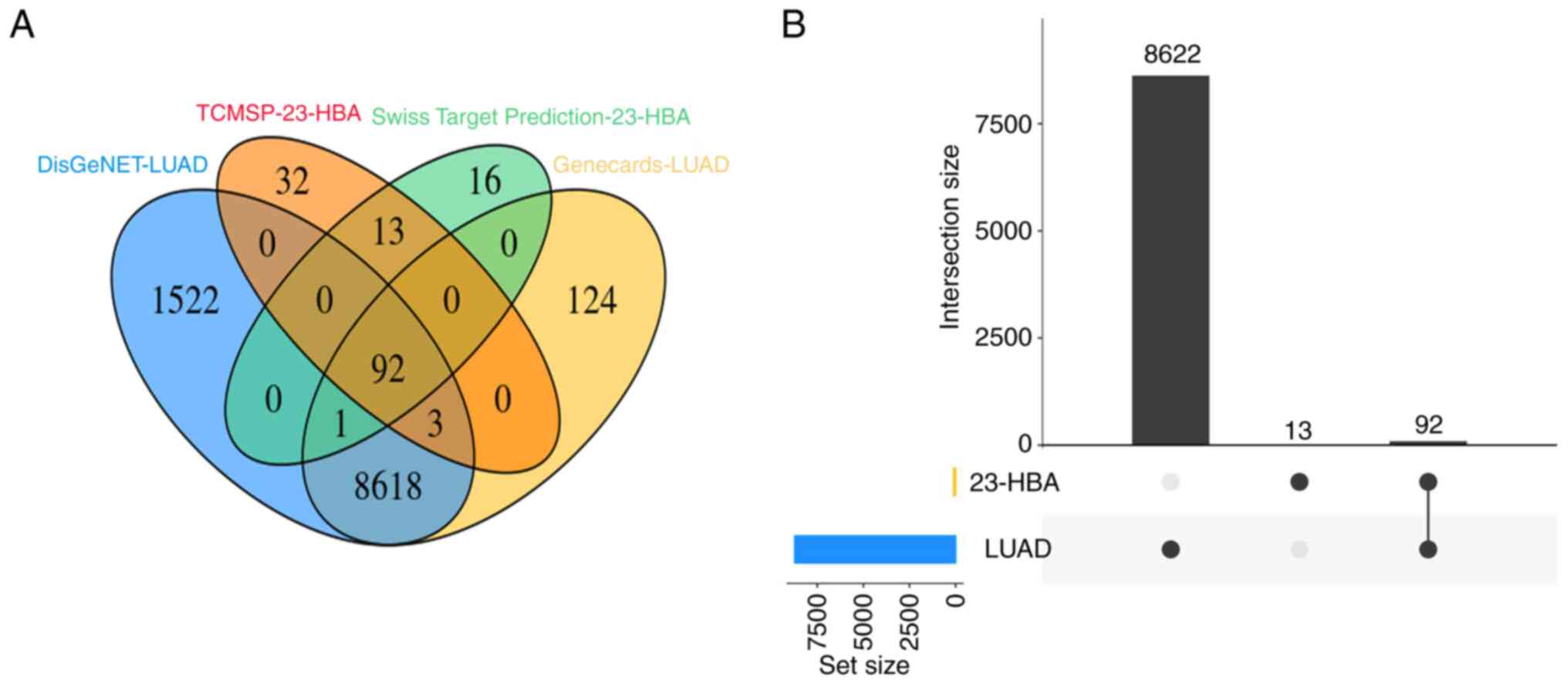
A total of >8,000 lung adenocarcinoma-related genes were screened through the DisGeNET (www.disgenet.org) and GeneCards (www.genecards.org) databases (28). Subsequently, the 23-HBA drug targets were identified through the Traditional Chinese Medicine Systems Pharmacology (TCMSP; old.tcmsp-e.com/tcmsp.p) and SwissTargetPrediction (www.swisstargetprediction.ch) databases. The 23-HBA potential targets and lung adenocarcinoma-related genes were mapped using Venny tool version 2.1.0 (bioinfogp.cnb.csic.es/tools/venny/). The total number of common targets was analyzed.
MTT assay for verification of the effect of 23-HBA on lung adenocarcinoma cells (29)
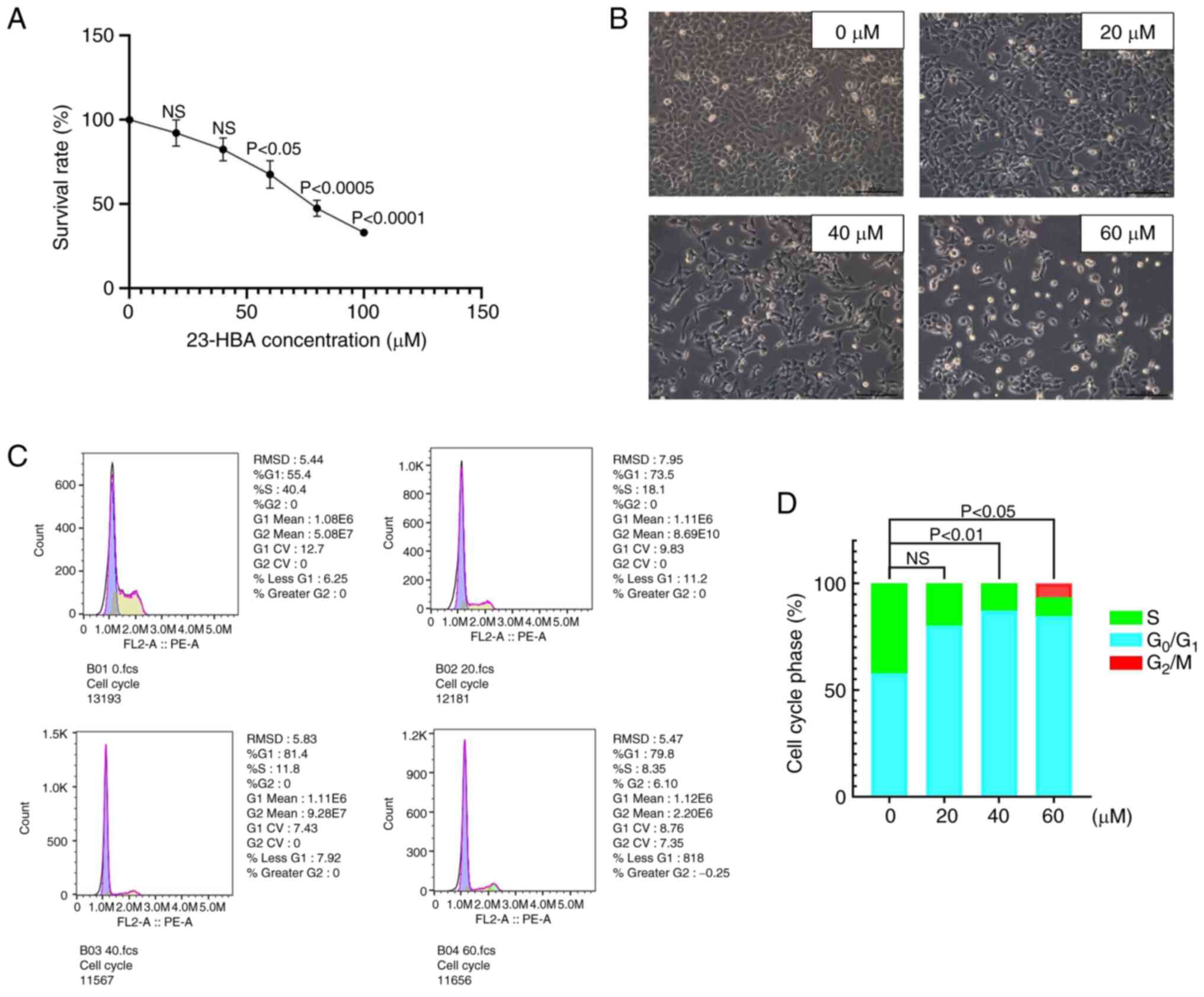
H1299 cells in the logarithmic growth phase were inoculated into 96-well plates, with 2,000 cells placed in each well. After 16-18 h, the cells completely attached to the wells, and were divided into two groups: Experimental group (drug group) and control group (treated with DMSO). Several drug doses (0, 40, 60, 80 and 100 µM) were examined to determine the optimal concentration for subsequent experiments. The cells were cultured with 23-HBA at 37˚C for 48 h. Subsequently, the supernatant was discarded, and 90 µl RPMI-1640 complete medium (Gibco; Thermo Fisher Scientific, Inc.) with 10 µl MTT solution were added to each well. The plates were incubated at 37˚C in the dark for 4 h, before the supernatant was discarded. Subsequently, 150 µl DMSO (Sigma Corporation) was added and gently mixed to dissolve the formazan crystals. An ELISA microplate detector (Biosharp Life Sciences) was used to measure the optical density values at a wavelength of 491 nm.
Cell cycle analysis by flow cytometry
H1299 cells in the logarithmic growth phase were inoculated at a concentration of 1x105 cells/well in a 6-well plate. After 16-18 h of culture, the cells were exposed to varying concentrations (0, 20, 40 and 60 µM) of 23-HBA. Following 48 h of treatment (37˚C), the supernatant was collected in a 2.0 ml Eppendorf tube. For cell collection, 300 µl trypsin (Sigma-Aldrich; Merck KGaA) was added to the cells and incubated at 37˚C for 3 min. When the cells became spherical in shape, an equivalent amount of RPMI-1640 complete was added to terminate digestion. The suspension was centrifuged (37˚C) at 300 x g, and the pellet was collected after 5 min. The single-cell suspension was washed with PBS (OriGene Technologies, Inc.) and then subjected to centrifugation under the same conditions. Subsequently, the suspension was fixed (4˚C) in 75% pre-cooled ethanol (500 µl) for a duration of 2 h to overnight (12 h), and was stored at 4˚C. Prior to staining, the fixed cells were washed with PBS. RNase A solution (100 µl) was added to the cell pellet, and the cells were incubated in a water bath at 37˚C for 30 min. Subsequently, 400 µl propidium iodide (Sigma-Aldrich; Merck KGaA) staining solution was added and thoroughly mixed, and the samples were incubated in the dark at 4˚C for 30 min. The cell cycle was determined using a flow cytometer (BD Biosciences) (30) and analyzed using FlowJo version 10.6.2 (Tree Star).
Protein-protein interaction (PPI) network
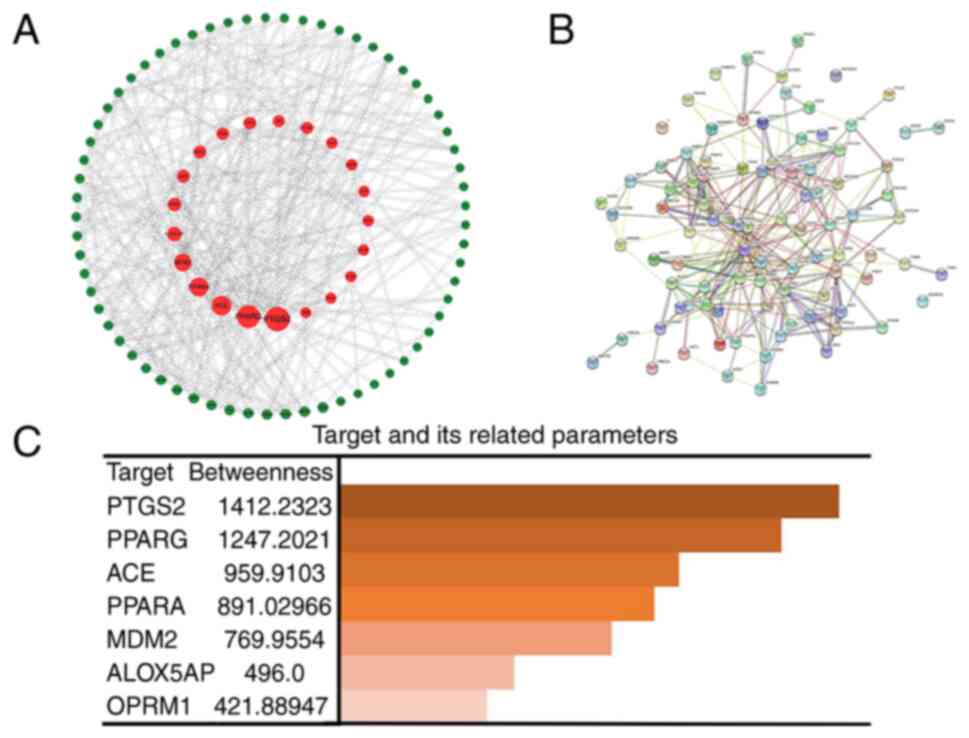
The targets associated with the pathogenesis of lung adenocarcinoma and the 23-HBA drug targets were imported into the STRING database (https://string-db.org/) to generate the PPI network of overlapping targets. Subsequently, the overlapping targets were imported into Cytoscape software version is 3.3 (https://cytoscape.org/) for visual analysis. Five core targets with a high degree of association between 23-HBA and lung adenocarcinoma were identified using the betweenness centrality (BC; the degree to which nodes are independent of each other) size sequencing (31), and the characteristics were analyzed. The characteristic is mainly manifested in: ACE mainly exists in vascular endothelial cells, while tumor cells have no blood vessels. Research on PTSG2 has been relatively clear and specific. MDM2 is highly expressed in tumors and has been proven to bind to p53. H1299 cells have a homozygous partial deletion of TP53 and therefore cannot express p53, and PPAR- and PPAR-γ belong to the same category and need not be studied. The median degree of freedom of the targets was 7.055, and the maximum degree of freedom was 18. The inclusion criteria for screening core targets were set at 15 and 18, and the best five core targets were selected based on the relevant parameters of each target (32).
Gene Expression Profiling Interactive Analysis (GEPIA) database
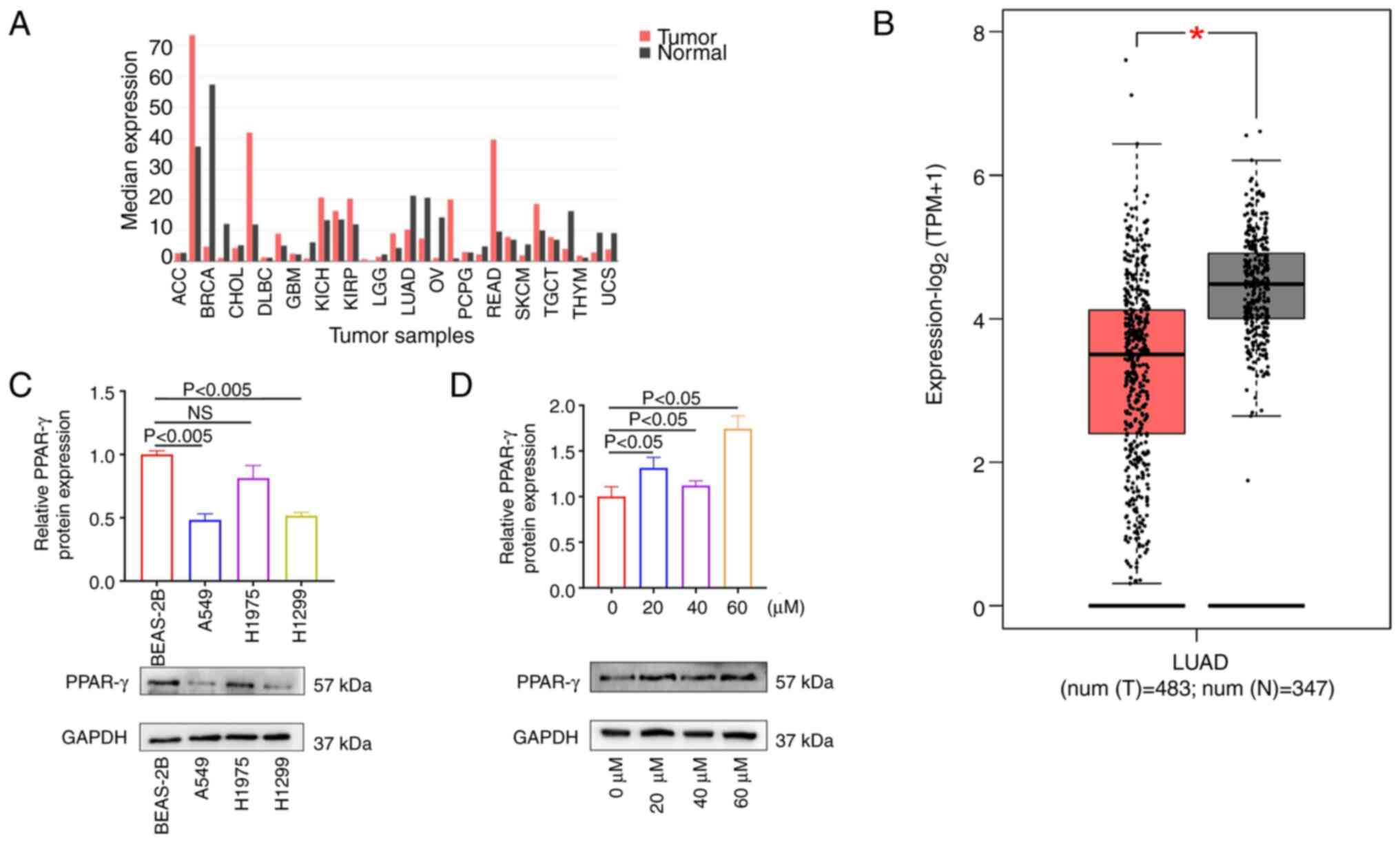
The mRNA expression profile of PPAR-γ in various tumor types, including lung adenocarcinoma, was assessed using the GEPIA database (http://gepia.cancer-pku.cn).
Western blot analysis
Western blot analysis was performed to compare the expression of PPAR-γ between normal and lung cancer cells and explore the characteristics of this protein (33). BEAS-2B, A549, H1975 and H1299 cells with robust growth profiles were collected. These cells were digested with trypsin, and the cell concentration was adjusted to 1x106 cells/ml. Subsequently, the cells were inoculated into a 6-well plate (1x106 cells per well) and cultured overnight. After a further 48-h incubation, the cells were digested with trypsin, and the cell precipitate was collected through centrifugation (200 x g, 37˚C, 3 min). RIPA buffer (Beyotime Institute of Biotechnology) and PMSF (Beijing Solarbio Science & Technology Co., Ltd.) were used to lyse cells (30 min with a 10 min mixing interval), followed by centrifugation (300 x g, 37˚C, 10 min). The supernatant was collected, and 5X SDS (Xilong Scientific Co., Ltd.) solution was added to the supernatant at a 1:4 ratio. The mixture was boiled at 95˚C for 10 min and stored at -20˚C. The protein concentration was determined using the bicinchoninic acid method. A total of 40 µg protein/lane were separated using 5% SDS-PAGE transferred to PVDF. The membranes were incubated with primary anti-GAPDH (rabbit anti-human GAPDH; cat. no. GTX100118; 1:1,000-5,000 dilution; Bioworld Technology) and PPAR-γ antibodies (PPAR-γ rabbit polyAb; cat. no. HY-P80872; 5 µg/ml; Protein Technology) at 4˚C for 6 h, followed by secondary anti-rabbit antibodies (goat anti-rabbit IgG-HRP; cat. no. L27A9; 2 µg/ml; Bioworld Technology) at 25˚C for 30 min. Following washing with 10% SDS, the membranes were incubated with ECL reagent (Biosharp Life Sciences) for signal development. The absorbance values of each band were calculated using ImageJ software (version 64; National Institutes of Health). GAPDH served as the internal reference. The entire experiment was repeated three times. Subsequently, H1299 cells were selected for subsequent experiments, and 23-HBA with various concentrations (0, 20, 40 and 60 µM) was applied to the cells at 37˚C for 2 h to analyze the expression of PPAR-γ.
Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analysis
KEGG analysis was performed using the R (version 4.1.2) package ‘Clusterprofiler’ (https://bioconductor.org), and pathways with P<0.05 and an association with cancer were screened (34). This process resulted in the identification of 12 pathways, which were then visualized and further screened for pathways exhibiting significant enrichment. It was visualized by using KEGG analysis; a graph is automatically generated when a gene is imported into the website for visualization purposes. GO enrichment analysis was conducted using the R package ‘Clusterprofiler’, and P<0.05 was used as the level of statistical significance. The relevant biological processes (BPs), cellular components (CCs) and molecular functions (MFs) were analyzed based on their P-value. Processes displaying high enrichment were selected for subsequent analyses.
Further exploration of the PPAR pathway-associated mechanism (35)
The target genes enriched in this pathway were identified through databases, such as KEGG (https://www.kegg.jp), to explore the mechanism of the PPAR signaling pathway. The mechanism of the PPAR pathway and its association with reactive oxygen species (ROS) were investigated by searching sources, such as PubMed (https://www.pubmed.ncbi.nlm.nih.gov) and Web of Science (https://clarivate.com.cn/solutions/web-of-science/), to predict the mechanism associated with the PPAR signaling pathway.
Mitochondrial ROS detection
H1299 cells stably cultured for several generations were used and diluted to 8x104 cells/ml after digestion. Of this cell suspension, 1 ml was added to a 6-well plate in addition to 1 ml of complete culture medium. After observing that the cells were almost completely attached to the wall, the old culture medium and was removed and 2 ml of 0, 20, 40 or 60 µmol/l 23-HBA culture medium was added to each well and incubated for 48 h. A ROS detection reagent (Beyotime Institute of Biotechnology) was employed for mitochondrial ROS detection. Dichlorodihydrofluorescein diacetate (DCFH-DA) was diluted with serum-free medium at a ratio of 1:1,000 to achieve a final concentration of 10 µmol/l. After the cell culture medium was discarded, an appropriate volume of diluted DCFH-DA was added to adequately cover the cells. The cells were then incubated in a 37˚C incubator for 20 min, followed by washing three times with serum-free medium to fully eliminate the DCFH-DA that did not enter the cells. Subsequently, the cells were observed directly using a laser confocal microscope (excitation wavelength, 488 nm; emission wavelength, 525 nm; Shanghai Tianneng Life Science Co., Ltd.).
Clinical prognosis
The target genes enriched in the PPAR signaling pathway were obtained through the GEPIA database (36). Kaplan-Meier curve analysis was then used to examine the effect of these target genes on the survival rate of patients with lung adenocarcinoma, with an aim to predict the clinical prognosis of these patients in association with the expression levels of the PPAR pathway target genes.
Statistical analysis
Data are presented as the mean ± SD of three experimental repeats. R (version 4.1.2) were utilized for the statistical analysis and required in the present study. The primary prognostic value was assessed using Kaplan-Meier curve analysis and the log-rank test, as well as Cox regression analysis. Other statistical analyses were performed in GraphPad Prism 6 software (Dotmatics). Comparisons among multiple groups were performed using one-way ANOVA followed by Tukey's post-hoc test, while Student's t-test comparing the experimental group with the control group one by one was used for comparisons between two groups. P<0.05 was considered do indicate a statistically significant difference.
Results
Association of 23-HBA with lung adenocarcinoma
Initially, genes associated with the pathogenesis of lung adenocarcinoma in the DisGeNET and Genecards databases and 23-HBA target genes in the SwissTargetPrediction and TCMSP databases were examined. Subsequently, a Venn diagram was constructed, revealing 92 overlapping genes (Fig. 1A). Further analysis demonstrated that these overlapping genes constituted 87.6% of the genes associated with 23-HBA (Fig. 1B). This finding provides evidence that the target genes of 23-HBA are closely linked to the pathogenesis of lung adenocarcinoma.
Inhibitory effect of 23-HBA on H1299 lung adenocarcinoma cells
First, the H1299 cell line, which was selected due to being cost-effective and producing reliable results, was cultured for MTT assays. The cells were divided into two groups, the control group and the experimental group, and various drug concentrations (0, 20, 40, 60, 80 and 100 µM) were used to explore the IC50 value of 23-HBA. The cell survival rate exhibited a steady decline with increasing concentrations of 23-HBA (Fig. 2A). At a drug concentration of 60 µM, which is close to the IC50 value, the cell survival rate was 54.4%. This finding provided evidence that 23-HBA had an inhibitory effect on the viability of H1299 cells. Therefore, in the follow-up experiments, drug doses between 0-60 µM were considered to be representative of the effect of 23-HBA. Microscopic analysis (fluorescence microscope) after treatment of the cells with 0-60 µM 23-HBA revealed a significant decrease in the number of adherent cells with an increase in drug concentration, accompanied by an increase in the number of floating dying cells (Fig. 2B). Finally, changes in the cell cycle of H1299 cells treated with various concentrations of 23-HBA were analyzed using flow cytometry (Fig. 2C), and the distribution of cells in each phase of the cell cycle was quantified (Fig. 2D). The proportion of H1299 cells in the G1 phase notably increased with increasing concentrations of 23-HBA, and, apart from the 20 µm group, this difference was statistically significant compared with the control group. However, fewer cells were in G2 and S phases. This finding indicated that 23-HBA affects H1299 cells by arresting the cell cycle in the G1 phase. All these results collectively demonstrated that 23-HBA inhibited the proliferation of lung adenocarcinoma H1299 cells.
PPAR-γ is the optimal target for the action of 23-HBA on lung adenocarcinoma cells
Firstly, the genes from the Venn diagram were imported into Cytoscape software, and topological parameter diagrams were created on the basis of the BC association of each gene for preliminary gene selection (Fig. 3A). Second, the related genes with lung adenocarcinoma pathogenesis genes and targets of the 23-HBA-associated domain were imported into STRING software to generate a PPI network diagram (Fig. 3B), which allowed for the analysis of gene associations to identify closely acting genes and exclude marginal genes of the network. Finally, the five most relevant genes associated with lung adenocarcinoma pathogenesis genes and targets of the 23-HBA-associated domain, prostaglandin-endoperoxide synthase 2, PPAR-γ, angiotensin I-converting enzyme (ACE), PPAR-α and mouse double minute 2 (MDM2), were selected and sorted in accordance with their association (Fig. 3C). ACE is a zinc metallopeptidase that is widely distributed in endothelial and epithelial cells. It plays a crucial role in the renin-angiotensin-aldosterone system. Consequently, ACE is mainly found in vascular endothelial cells (37). For PTSG2, the related studies have been relatively clear and specific, so we excluded it from further analysis. MDM2, as a key negative regulator of p53, exhibits high expression in tumors and serves an important role in tumor initiation and development. MDM2 has been demonstrated to bind to p53, thereby blocking its tumor-suppressor transactivation domain (38). Furthermore, MDM2 functions as an E3 ligase, marking p53 for degradation by the proteasome. H1299 cells possess a homozygous partial deletion of TP53(39), rendering them unable to express the tumor-suppressor protein p53, which contributes to their proliferative phenotype. Therefore, MDM2 was excluded from further analysis. PPAR-α and PPAR-γ belong to the same category. PPAR-γ has been rarely studied in the literature and it represents a novel finding. Therefore, it was selected for exploration in the present study.
PPAR-γ functions as a tumor suppressor in lung adenocarcinoma
The GEPIA database was utilized to analyze the expression of the target gene PPAR-γ in various tumor types (40), including lung adenocarcinoma (Fig. 4A). The results demonstrated that PPAR-γ was significantly downregulated in lung adenocarcinoma compared with normal tissues (Fig. 4B), while being highly expressed in most tumors, thereby indicating that PPAR-γ may function as a tumor-suppressor gene. Western blot assay was performed to measure PPAR-γ expression in normal cells (BEAS-2B) and multiple lung adenocarcinoma cell lines (A549, H1975 and H1299), to further confirm its role as a tumor suppressor. The results showed that PPAR-γ was highly expressed in normal cells, whereas its expression was notably lower in the lung adenocarcinoma cell lines (Fig. 4C). Subsequently, western blotting was conducted to examine the protein expression of PPAR-γ after treatment of the cells with different concentrations of 23-HBA. The results indicated that the expression of PPAR-γ in the 23-HBA treatment groups was higher than that in the control group (41), with statistical significance observed at a drug concentration of 60 µM (Fig. 4D). These results confirmed that 23-HBA treatment increases the expression of PPAR-γ. Given the earlier conclusion that 23-HBA can inhibit the survival of lung adenocarcinoma cells, PPAR-γ may act as a tumor suppressor, which is consistent with the results from the GEPIA database.
Mitochondrial ROS is associated with the mechanism of inhibition of lung adenocarcinoma cell proliferation by 23-HBA
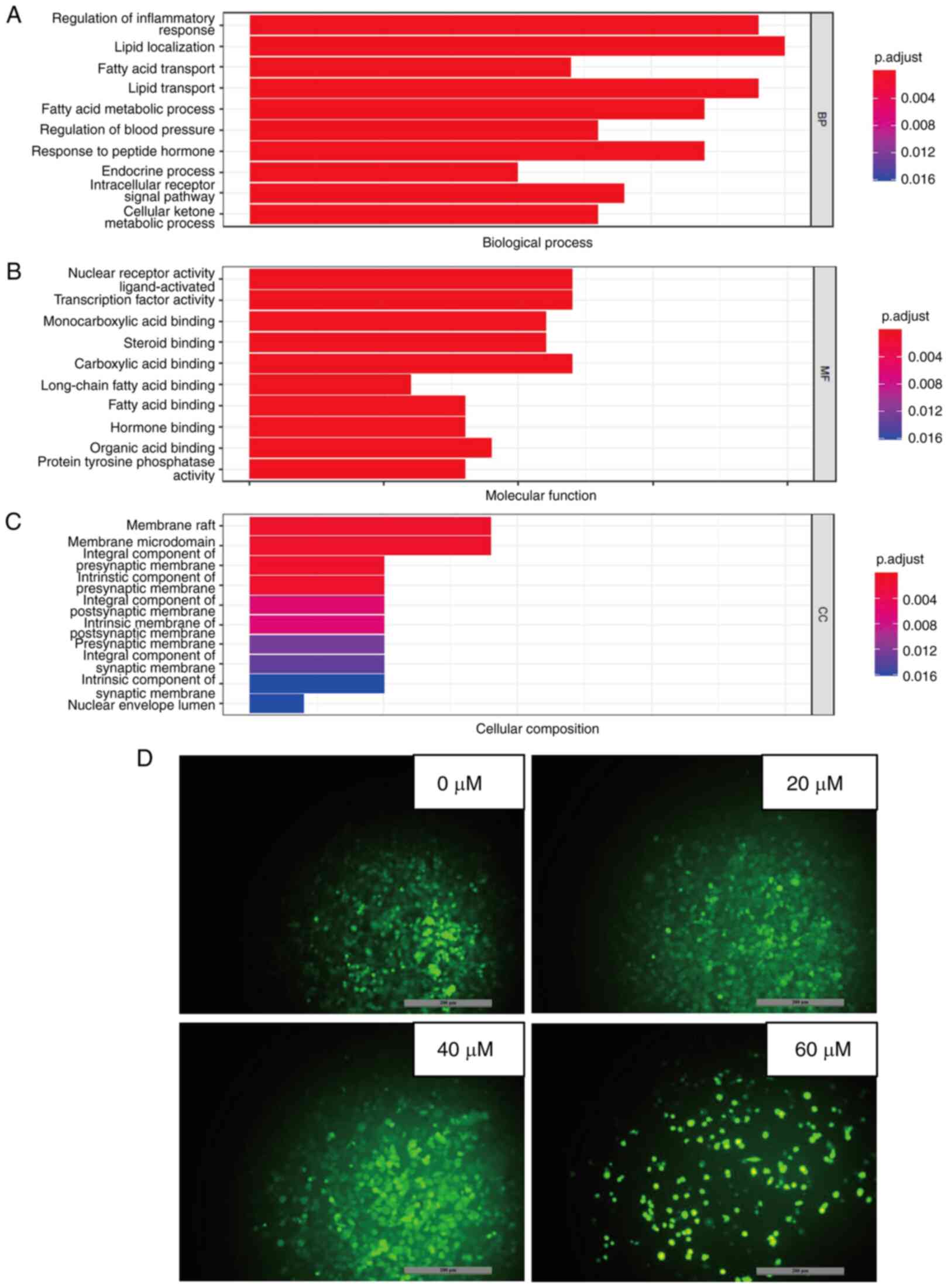
GO analysis was conducted to investigate the mode of action of 23-HBA on lung adenocarcinoma (42). P<0.05 was used as the screening criterion for selected functional terms in the categories BP, MF and CC (Fig. 5A-C). The results showed that most of the enriched BP terms require the consumption of ATP, which is primarily produced by mitochondria. From Fig. 5A, it can be concluded that most enriched BP terms are associated with regulation of inflammatory response, which require the consumption of ATP, such as lipid transporting and fatty acid metabolic processing. The most important mechanisms of action in the category MF are associated with nuclear receptor activity, which also contains ligand-activated transcription factor activity and carboxylic acid binding. The most dominant mechanism of action in the category CC is membrane raft, which also contains membrane microdomain and integral component of presynaptic membrane. Therefore, the mechanism of action of the PPAR pathway is related to mitochondria due to increasing the content of mitochondrial ROS, thereby inhibiting cell growth. Further investigation through PubMed into the PPAR signaling pathway revealed that it induces physiological changes, including inflammation and alterations in the mitochondrial membrane potential (43). Therefore, the mechanism by which 23-HBA inhibits the viability and/or proliferation of lung adenocarcinoma cells may be related to mitochondrial ROS. Subsequently, the mitochondrial ROS levels were detected, and the results showed that the production of intracellular mitochondrial ROS notably increased with increasing concentrations of 23-HBA (Fig. 5D), which is consistent with the aforementioned mechanism.
Clinical prognosis of patients with lung adenocarcinoma is associated with the levels of PPAR signaling pathway-related genes
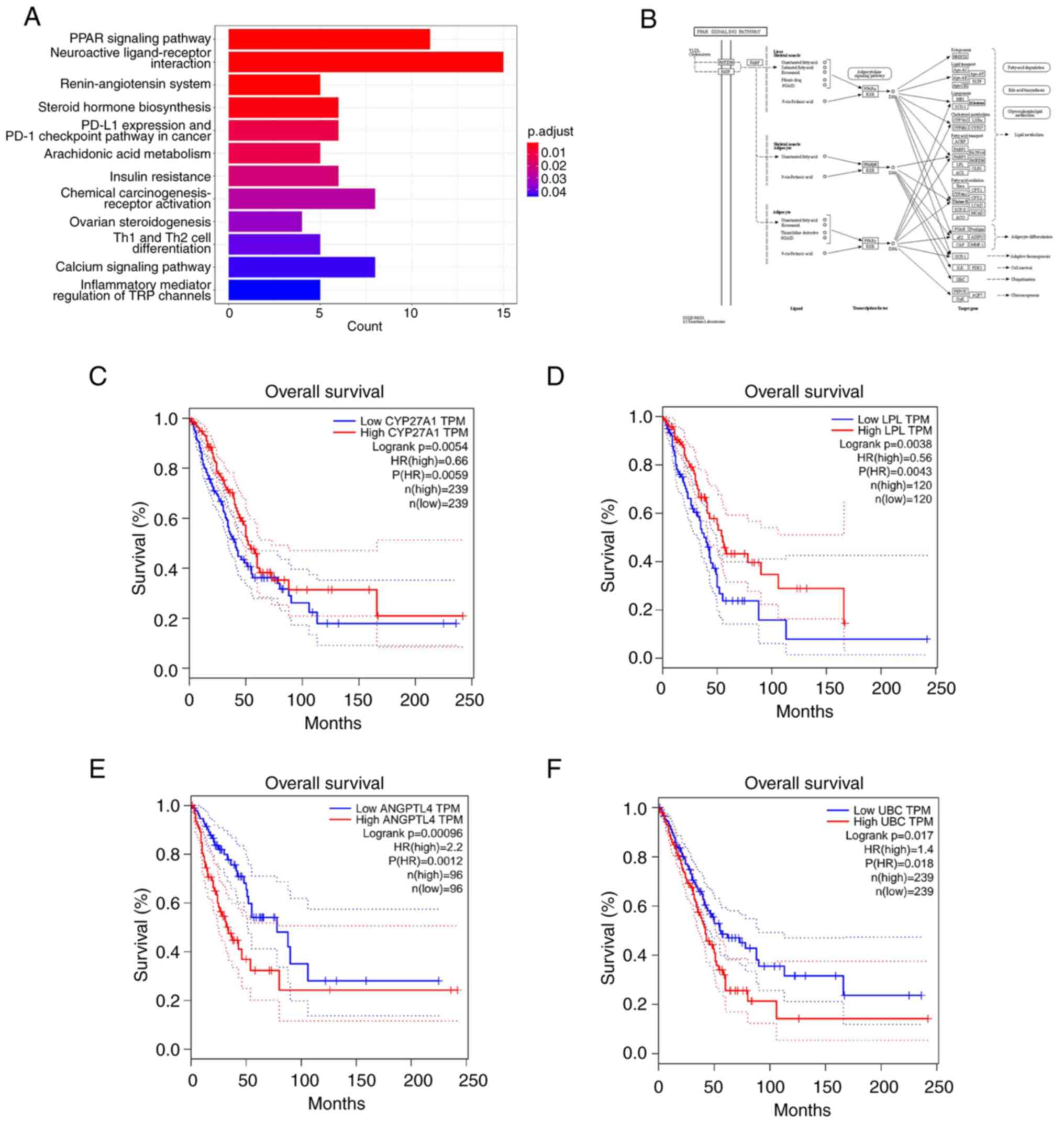
The PPAR signaling pathway is considered to be an important signal transduction pathway in several conditions, such as inflammation and cancer (44). Previous studies have demonstrated the inhibitory effects of 23-HBA and its derivatives on inflammation, as well as liver, lung and breast cancer (45,46). The KEGG analysis revealed that the pathway exhibiting the highest degree of enrichment was for the ‘PPAR signaling pathway’ (Fig. 6A). Investigation of the research mechanism within this pathway resulted in the identification of target genes associated with it, including CYP27, LPL, PGAR/ANGPTL4 and UBC (Fig. 6B). Subsequently, the GEPIA database was used to analyze the association of the expression levels of these target genes with the survival of patients with lung adenocarcinoma. The results indicated that high (ANGPTL4 and UBC) or low (CYP27 and LPL) expression levels of these genes were significantly associated with a lower patient survival (Fig. 6C-F), thereby demonstrating the clinical prognostic significance of the PPAR pathway.
Effect of 23-HBA on lung adenocarcinoma and its mechanism of action
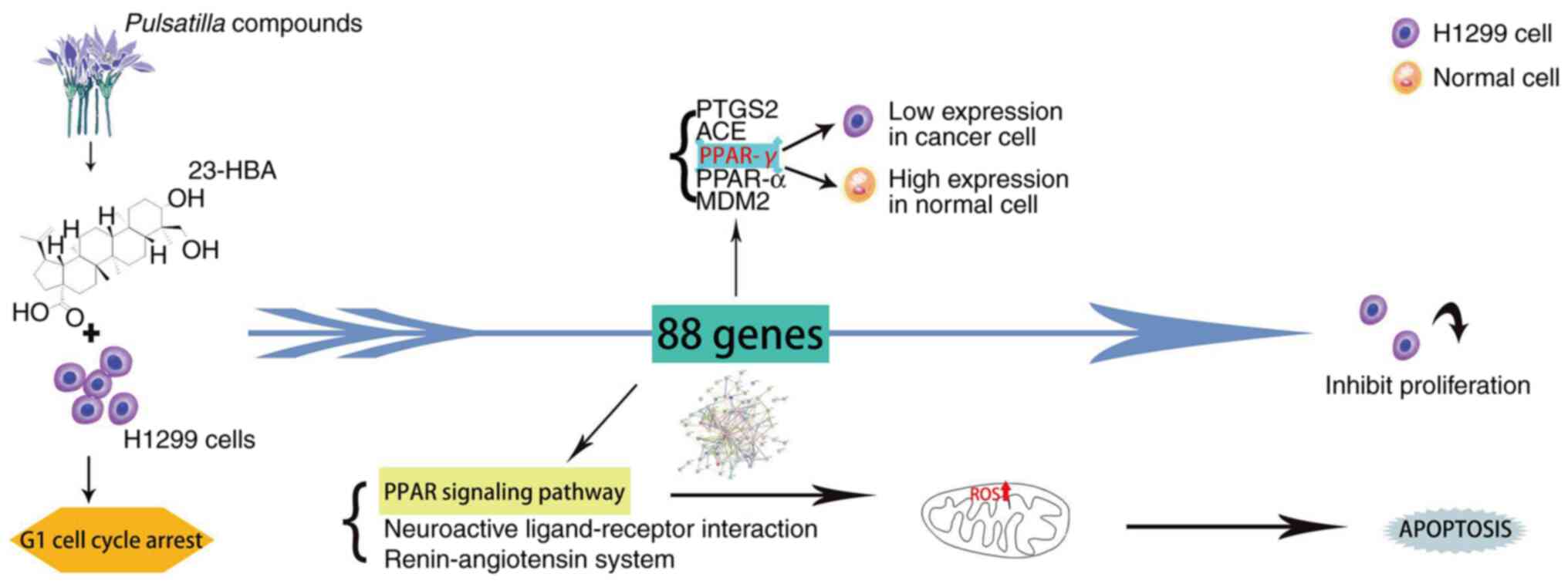
Collectively, the results of the present study demonstrated that 23-HBA inhibits cell proliferation by blocking lung adenocarcinoma cells in the G1 phase. The results indicated that PPAR-γ may function as a tumor-suppressor gene in lung adenocarcinoma. Further analysis unveiled the relationship among this finding, the PPAR signaling pathway and mitochondrial ROS. The relationship is that the survival rate of tumor cells decreases with the increase of drug concentration, and at the same time, the expression of ROS increases with the increase of drug concentration. Finally, a clinical prognosis association was demonstrated between the expression levels of PPAR pathway-related genes and the survival of patients with lung adenocarcinoma (Fig. 7).
Discussion
PPAR-γ is a nuclear receptor involved in various cellular processes, including regulation of gene expression that serves an important regulatory function in the process of lipid metabolism (47). Studies have indicated that PPAR-γ plays a pivotal role in antitumor mechanisms by governing cell cycle arrest, apoptosis, proliferation, invasion and migration, among other functions (48,49). Western blot analysis in the present study indicated that PPAR-γ functions as a tumor-suppressor gene by inhibiting the growth of lung adenocarcinoma cells. In a previous study, PPAR-γ gene silencing and overexpression vectors were constructed for the study of human squamous cell carcinoma. These vectors were transfected into human squamous cell carcinoma Colo16 cells to assess the effects of PPAR-γ on cell proliferation and apoptosis. The results indicated that PPAR-γ silencing promoted Colo16 cell proliferation and reduced apoptosis, whereas PPAR-γ overexpression exhibited the opposite effects. Therefore, PPAR-γ serves a role in inhibiting tumor cell proliferation and promoting apoptosis in human squamous cell carcinoma Colo16 cells (50). The conclusions of the present study align with these findings. However, no specific mechanism concerning the involvement of PPAR-γ-associated genes in lung adenocarcinoma has been reported. In the present study, PPAR-γ was shown, to be downregulated in lung adenocarcinoma cells and function as a tumor-suppressor gene (51,52).
KEGG analysis was conducted on the selected intersecting genes to delve further into the mechanism of 23-HBA in lung adenocarcinoma (20). The pathway exhibiting the highest degree of enrichment was the ‘PPAR signaling pathway’, which plays a pivotal regulatory role in immunity (53), inflammation, vascular function (54), cell proliferation, differentiation, development, apoptosis and cancer. This pathway is not only implicated in immune function, carbohydrate and lipid metabolism, but is also closely associated with the proliferation, death, invasion and metastasis of tumor cells during the course of tumor development. Activation of the PPAR pathway affects angiogenesis and tumor suppression, and regulates the proliferation and differentiation of cancer cells (55).
Previous studies reported that Pulsatilla compounds play a crucial role in inhibiting the growth of tumor cells, particularly in liver, lung and breast cancer (56,57). B4G6, a derivative of 23-HBA, has been demonstrated to induce mitochondrial apoptosis by triggering ROS accumulation and the opening of calcium-dependent permeability transition pores, thereby inhibiting the proliferation of HepG2, Hep3B, Bel-7402 and HepG2/ADM cell lines (58). B4G2 exhibits a potent inhibitory effect on HepG2 cell proliferation and induces mitochondrial autophagy. Furthermore, the use of autophagy inhibitors could enhance the apoptosis rate of HepG2 cells (59). Experiments involving mitochondrial ROS were conducted to further explore the mechanism of 23-HBA in lung adenocarcinoma. The results showed that the accumulation of intracellular mitochondrial ROS significantly increased with an increase in drug concentration.
Furthermore, MTT and flow cytometry analyses were employed in the present study to assess the effect of 23-HBA on lung adenocarcinoma cells (60). The MTT results indicated that 23-HBA had an inhibitory effect on lung adenocarcinoma cell survival. Moreover, 23-HBA caused the arrest of H1299 cells in the G1 cell cycle phase, as determined by flow cytometry. In summary, 23-HBA was shown to effectively inhibit the survival and proliferation of lung adenocarcinoma cells. This finding establishes a theoretical foundation for the development and clinical application of new antitumor drugs based on Pulsatilla compounds (61). Developing high-efficiency and low-toxicity drugs with Pulsatilla compounds as the primary constituents may aid in inhibiting tumor growth in the future (62). In this context, 23-HBA may represent a promising therapeutic strategy for lung adenocarcinoma by interacting with the PPAR-γ and mitochondrial ROS pathways (63).
In summary 23-HBA was shown to inhibit cell proliferation by arresting lung adenocarcinoma cells in the G1 phase, and PPAR-γ was indicated to be a tumor-suppressor gene in lung adenocarcinoma. In addition, the PPAR signaling pathway had the highest degree of enrichment among genes associated with 23-HBA and lung adenocarcinoma; however, the mechanism involved was not clarified, and further animal experiments are required to elucidate the inhibitory effects of 23-HBA on lung adenocarcinoma in vivo. 23-HBA is the active ingredient extracted from the Radix Pulsatilla, a plant used in traditional Chinese medicine. However, the use of traditional Chinese medicine in cancer treatment is uncommon. Therefore, the present study could set the foundation for the potential application of traditional Chinese medicine in lung adenocarcinoma treatment, thereby expanding new avenues for the development of traditional Chinese and Western medicines.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Shandong Science and Technology Committee (grant no. ZR2023MH223), the National Natural Science Foundation of China (grant no. 81800169) and College Students' Innovation and Entrepreneurship Training Program (grant nos. 202310440061, 202310440029 and S202310440012).
Availability of data and materials
The data generated in the present study may be requested from the corresponding author.
Authors' contributions
BT designed and performed the cell experiments. XL screened the final drug through cell experiments. YZ performed the database search and data processing, and assisted in the experimental design. PL and QJ analyzed the data. ZW, ZL, WS and YX assisted in the cell experiments. YL and YS designed the experiments and revised the manuscript. All authors read and approved the final manuscript. All authors confirm the authenticity of all the raw data.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A and Bray F: Gloal cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.PubMed/NCBI View Article : Google Scholar | |
|
Shigeta K, Datta M, Hato T, Kitahara S, Chen IX, Matsui A, Kikuchi H, Mamessier E, Aoki S, Ramjiawan RR, et al: Dual programmed death receptor-1 and vascular endothelial growth factor receptor-2 blockade promotes vascular normalization and enhances antitumor immune responses in hepatocellular carcinoma. Hepatology. 71:1247–1261. 2020.PubMed/NCBI View Article : Google Scholar | |
|
Aoki T, Kudo M, Ueshima K, Morita M, Chishina H, Takita M, Hagiwara S, Ida H, Minami Y, Tsurusaki M and Nishida N: Incidence of hyper progressive disease in combination immunotherapy and anti-programmed cell death protein 1/programmed death-ligand 1 monotherapy for unresectable hepatocellular carcinoma. Liver Cancer. 13:56–69. 2023.PubMed/NCBI View Article : Google Scholar | |
|
Cheng X: The effect of CDKN2A gene mutation on lung cancer and its relationship with prognosis. J Appl Cancer. 875-878(883)2021. | |
|
Oudkerk M, Liu S, Heuvelmans MA, Walter JE and Field JK: Lung cancer LDCT screening and mortality reduction-evidence, pitfalls and future perspectives. Nat Rev Clin Oncol. 18:135–151. 2021.PubMed/NCBI View Article : Google Scholar | |
|
Zhang L, Shao F, Li L, et al: CALM1 gene expression and EGFR mutation in colorectal cancer and its clinical significance. Gen Med Edu. 21:18–21. 2023.DOI: 10.13558/j. carol carroll nki issn1672-3686.2023.001.005. | |
|
Brown A and Goodman Z: Hepatitis B-associated fibrosis and fibrosis/cirrhosis regression with nucleoside and nucleotide analogs. Expert Rev Gastroenterol Hepatol. 6:187–198. 2012.PubMed/NCBI View Article : Google Scholar | |
|
Clough E and Barrett T: The gene expression omnibus database. Methods Mol Biol. 1418:93–110. 2016.PubMed/NCBI View Article : Google Scholar | |
|
Huang J, Zhang J, Zhang F, Lu S, Guo S, Shi R, Zhai Y, Gao Y, Tao X, Jin Z, et al: Identification of a disulfidptosis-related genes signature for prognostic implication in lung adenocarcinoma. Comput Biol Med. 165(107402)2023.PubMed/NCBI View Article : Google Scholar | |
|
Liang Q, Gong M, Zou JH, Luo MY, Jiang LL, Wang C, Shen NX, Zhang MC, Xu L, Lei HM, et al: A phosphoglycerate mutase 1 allosteric inhibitor overcomes drug resistance to EGFR-targeted therapy via disrupting IL-6/JAK2/STAT3 signaling pathway in lung adenocarcinoma. Drug Resist Updat. 68(100957)2023.PubMed/NCBI View Article : Google Scholar | |
|
Jiang M, Yang M, Zhou YY, et al: Effects of 23-hydroxybetulinic acid on proliferation and apoptosis of vascular endothelial cells. Cancer Prev Res. 911–913+985. 2007. | |
|
Molina JR, Yang P, Cassivi SD, Schild SE and Adjei AA: Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594. 2008.PubMed/NCBI View Article : Google Scholar | |
|
Tang Z, Kang B, Li C, Chen T and Zhang Z: GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47 (W1):W556–W560. 2019.PubMed/NCBI View Article : Google Scholar | |
|
Caligiuri G: CD31 as a therapeutic target in atherosclerosis. Circ Res. 126:1178–1189. 2020.PubMed/NCBI View Article : Google Scholar | |
|
Liao D, Sundlov J, Zhu J, Mei H, Hu Y, Newman DK and Newman PJ: Atomic level dissection of the platelet endothelial cell adhesion molecule 1 (PECAM-1) homophilic binding interface: Implications for endothelial cell barrier function. Arterioscler Thromb Vasc Biol. 42:193–204. 2022.PubMed/NCBI View Article : Google Scholar | |
|
Hu M, Zhang H, Liu Q and Hao Q: Structural basis for human PECAM-1-mediated trans-homophilic cell adhesion. Sci Rep. 6(38655)2016.PubMed/NCBI View Article : Google Scholar | |
|
Winneberger J, Schöls S, Lessmann K, Rández-Garbayo J, Bauer AT, Mohamud Yusuf A, Hermann DM, Gunzer M, Schneider SW, Fiehler J, et al: Platelet endothelial cell adhesion molecule-1 is a gatekeeper of neutrophil transendothelial migration in ischemic stroke. Brain Behav Immun. 93:277–287. 2021.PubMed/NCBI View Article : Google Scholar | |
|
Zhu H, Lu L, Zhu W, Tan Y, Duan Y, Liu J, Ye W, Zhu Z, Xu J and Xu S: Design and synthesis of NAD(P)H: Quinone oxidoreductase (NQO1)-activated prodrugs of 23-hydroxybetulinic acid with enhanced antitumor properties. Eur J Med Chem. 240(114575)2022.PubMed/NCBI View Article : Google Scholar | |
|
Chen SY, Tsuneyama K, Yen MH, Lee JT, Chen JL and Huang SM: Hyperbaric oxygen suppressed tumor progression through the improvement of tumor hypoxia and induction of tumor apoptosis in A549-cell-transferred lung cancer. Sci Rep. 11(12033)2021.PubMed/NCBI View Article : Google Scholar | |
|
Tian D, Yu Y, Zhang L, Sun J and Jiang W: 23-Hydroxybetulinic acid reduces tumorigenesis, metastasis and immunosuppression in a mouse model of hepatocellular carcinoma via disruption of the MAPK signaling pathway. Anticancer Drugs. 33:815–825. 2022.PubMed/NCBI View Article : Google Scholar | |
|
Ye B and Ji ZN: 23-Hydroxybetulinic acid-induced HL-60 cell autophagic apoptosis and its molecular mechanism. Nat Prod Res. 26:1063–1068. 2012.PubMed/NCBI View Article : Google Scholar | |
|
Doi H, Kida T, Nishino K, Nakatsuji M, Sakamoto S, Shimizu S, Teraoka Y, Tamura Y, Kataoka Y and Inui T: Solubility-improved 10-O-substituted SN-38 derivatives with antitumor activity. ChemMedChem. 12:1715–1722. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Xiang S: Effect and mechanism of 23-hydroxybetulcholic acid on ulcerative colitis induced by DSS. Zunyi Medical Coll, 2022. DOI: 10.27680 /, dc nki. Gzyyc. 2022.000496. | |
|
Wang C, Yu Q, Song T, Wang Z, Song L, Yang Y, Shao J, Li J, Ni Y, Chao N, et al: The heterogeneous immune landscape between lung adenocarcinoma and squamous carcinoma revealed by single-cell RNA sequencing. Signal Transduct Target Ther. 7(289)2022.PubMed/NCBI View Article : Google Scholar | |
|
Kobayashi Y and Mitsudomi T: Not all epidermal growth factor receptor mutations in lung cancer are created equal: Perspectives for individualized treatment strategy. Cancer Sci. 107:1179–1186. 2016.PubMed/NCBI View Article : Google Scholar | |
|
Li Q, Zhang H, Yan X, Zhao Z, Qiu J, Hu L, Jiang S, Kong Q, Sun J and Li L: Up-regulation of PPAR-γ involved in the therapeutic effect of icariin on cigarette smoke-induced inflammation. Pulm Pharmacol Ther. 79(102197)2023.PubMed/NCBI View Article : Google Scholar | |
|
Chen JJ, Patel A, Sodani K, Xiao ZJ, Tiwari AK, Zhang DM, Li YJ, Yang DH, Ye WC, Chen SD and Chen ZS: BBA, a synthetic derivative of 23-hydroxybutulinic acid, reverses multidrug resistance by inhibiting the efflux activity of MRP7 (ABCC10). PLoS One. 8(e74573)2013.PubMed/NCBI View Article : Google Scholar | |
|
Chen H, Bai L, Shi Y, Zhang X, Wang X, Wang Y, Hu J and Zhou P: Investigation of the molecular mechanism underlying the therapeutic effect of perilla frutescens L. Essential oil on acute lung injury using gas chromatography mass spectrometry and network pharmacology. Comb Chem High Throughput Screen: Oct 10, 2023 (Epub ahead of print). | |
|
Huang Z, Gao Y and Hou D: Interleukin-22 enhances chemoresistance of lung adenocarcinoma cells to paclitaxel. Hum Cell. 33:850–858. 2020.PubMed/NCBI View Article : Google Scholar | |
|
Feng Z, Chen Y, Cai C, Tan J, Liu P, Chen Y, Shen H, Zeng S and Han Y: Pan-cancer and single-cell analysis reveals CENPL as a cancer prognosis and immune infiltration-related biomarker. Front Immunol. 13(916594)2022.PubMed/NCBI View Article : Google Scholar | |
|
Cao J, Sun S, Min R, Li R, Fan X, Han Y, Feng Z and Li N: Prognostic significance of CCNB2 expression in triple-negative breast cancer. Cancer Manag Res. 13:9477–9487. 2021.PubMed/NCBI View Article : Google Scholar | |
|
Zhang W, Tian W, Wang Y, Jin X, Guo H, Wang Y, Tang Y and Yao X: Explore the mechanism and substance basis of Mahuang FuziXixin Decoction for the treatment of lung cancer based on network pharmacology and molecular docking. Comput Biol Med. 151(106293)2022.PubMed/NCBI View Article : Google Scholar | |
|
Du Y, Lv Z, Sun D, Li Y, Sun L and Zhou J: Physcion 8-O-β-glucopyranoside exerts anti-tumor activity against non-small cell lung cancer by targeting PPARγ. Anat Rec (Hoboken). 302:785–793. 2019.PubMed/NCBI View Article : Google Scholar | |
|
Kim S and Bae S: In vitro and in vivo human body odor analysis method using GO:PANI/ZNRs/ZIF-8 adsorbent followed by GC/MS. Molecules. 27(4795)2022.PubMed/NCBI View Article : Google Scholar | |
|
Lei Y: Anti-liver cancer activity evaluation and mechanism of 23-hydroxybetulinic acid derivative B4G2. Jinan University, 2017. | |
|
Ma J, Cai X, Kang L, Chen S and Liu H: Identification of novel biomarkers and candidate small-molecule drugs in cutaneous melanoma by comprehensive gene microarrays analysis. J Cancer. 12:1307–1317. 2021.PubMed/NCBI View Article : Google Scholar | |
|
Liu L, Zhou S, Yang Z, et al: Study on population distribution of ACE gene polymorphism and its high expression in essential hypertension. J Pract Lab Phys. 10:132–134. 2018. | |
|
Ding H and Hu M: Research progress of anti-tumor small molecule inhibitors targeting p53-MDM2. Adv Clin Med. 13:13474–13483. 2023.(In Chinese). | |
|
Xiong Y and Li B: The cause of deletion of P53 protein in H1299 cells. Macau Med J. 4:8–9. 2004. | |
|
Dourado KMC, Baik J, Oliveira VKP, Beltrame M, Yamamoto A, Theuer CP, Figueiredo CAV, Verneris MR and Perlingeiro RCR: Endoglin: A novel target for therapeutic intervention in acute leukemias revealed in xenograft mouse models. Blood. 129:2526–2536. 2017.PubMed/NCBI View Article : Google Scholar | |
|
liu xiao mei, first hai-li wang, etc: Research progress on the role of PPAR-γ/AP-1 signaling pathway in related diseases. J Yichun Univ, 21,43: 12-16. | |
|
Moloney JN and Cotter TG: ROS signalling in the biology of cancer. Semin Cell Dev Biol. 80:50–64. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Cao XJ, Zhang MJ, Zhang LL, Yu K, Xiang Y, Ding X, Fan J, Li JC and Wang QS: TLR4 mediates high-fat diet induced physiological changes in mice via attenuating PPARγ/ABCG1 signaling pathway. Biochem Biophys Res Commun. 503:1356–1363. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Alqahtani S and Mahmoud AM: Gamma-Glutamylcysteine Ethyl Ester Protects against Cyclophosphamide-Induced Liver Injury and Hematologic Alterations via Upregulation of PPAR and Attenuation of Oxidative Stress, Inflammation, and Apoptosis. Oxidative Medicine and Cellular Longevity, 2016. | |
|
Laganà AS, Vitale SG, Nigro A, Sofo V, Salmeri FM, Rossetti P, Rapisarda AM, La Vignera S, Condorelli RA, Rizzo G and Buscema M: Pleiotropic actions of peroxisome proliferator-activated receptors (PPARs) in dysregulated metabolic homeostasis, inflammation and cancer: Current evidence and future perspectives. Int J Mol Sci. 17(999)2016.PubMed/NCBI View Article : Google Scholar | |
|
Jiang MJ, Yang M, Zhou YY, Zhang RJ, Cao GX, Cai GM and Wang GJ: In vitro inhibitory effect of 23-HBA on angiogenesis. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 22:88–91. 2006.PubMed/NCBI(In Chinese). | |
|
Zhang DM, Shu C, Chen JJ, Sodani K, Wang J, Bhatnagar J, Lan P, Ruan ZX, Xiao ZJ, Ambudkar SV, et al: BBA, a derivative of 23-hydroxybetulinic acid, potently reverses ABCB1-mediated drug resistance in vitro and in vivo. Mol Pharm. 9:3147–3159. 2012.PubMed/NCBI View Article : Google Scholar | |
|
Nahlé N: PPAR trilogy from metabolism to cancer. Curr Opin Clin Nutr Metab Care. 7:397–402. 2004.PubMed/NCBI View Article : Google Scholar | |
|
Elix CC, Salgia MM, Otto-Duessel M, Copeland BT, Yoo C, Lee M, Tew BY, Ann D, Pal SK and Jones JO: Peroxisome proliferator-activated receptor gamma controls prostate cancer cell growth through AR-dependent and independent mechanisms. Prostate. 80:162–172. 2020.PubMed/NCBI View Article : Google Scholar | |
|
Zhang J and Bai WP: C1q/tumor necrosis factor related protein 6 (CTRP6) regulates the phenotypes of high glucose-induced gestational trophoblast cells via peroxisome proliferator-activated receptor gamma (PPARγ) signaling. Bioengineered. 13:206–216. 2022.PubMed/NCBI View Article : Google Scholar | |
|
Zhou P: Preliminary study on the function of PPARG gene in human skin squamous cell carcinoma Colol6 cells. Peking Union Medical College, 2019. | |
|
Zhan L, Zhang H, Zhang Q, Woods CG, Chen Y, Xue P, Dong J, Tokar EJ, Xu Y, Hou Y, et al: Regulatory role of KEAP1 and NRF2 in PPARγ expression and chemoresistance in human non-small-cell lung carcinoma cells. Free Radic Biol Med. 53:758–768. 2012.PubMed/NCBI View Article : Google Scholar | |
|
Li H, Sorenson AL, Poczobutt J, Amin J, Joyal T, Sullivan T, Crossno JT Jr, Weiser-Evans MC and Nemenoff RA: Activation of PPARgamma in myeloid cells promotes lung cancer progression and metastasis. PLoS One. 6(e28133)2011.PubMed/NCBI View Article : Google Scholar | |
|
Liu S, Zhang H, Li Y, Zhang Y, Bian Y, Zeng Y, Yao X, Wan J, Chen X, Li J, et al: S100A4 enhances protumor macrophage polarization by control of PPAR-γ-dependent induction of fatty acid oxidation. J Immunother Cancer. 9(e002548)2021.PubMed/NCBI View Article : Google Scholar | |
|
Wagner N and Wagner KD: Pharmacological utility of PPAR modulation for angiogenesis in cardiovascular disease. Int J Mol Sci. 24(2345)2023.PubMed/NCBI View Article : Google Scholar | |
|
Fang C, Liu Y, Chen L, Luo Y, Cui Y, Zhang N, Liu P, Zhou M and Xie Y: α-Hederin inhibits the growth of lung cancer A549 cells in vitro and in vivo by decreasing SIRT6 dependent glycolysis. Pharm Biol. 59:11–20. 2021.PubMed/NCBI View Article : Google Scholar | |
|
Reka AK, Goswami MT, Krishnapuram R, Standiford TJ and Keshamouni VG: Molecular cross-regulation between PPAR-γ and other signaling pathways: Implications for lung cancer therapy. Lung Cancer. 72:154–159. 2011.PubMed/NCBI View Article : Google Scholar | |
|
Łaska G, Sieniawska E, Maciejewska-Turska M, Świątek Ł, Pasco DS and Balachandran P: Pulsatilla vulgaris inhibits cancer proliferation in signaling pathways of 12 reporter genes. Int J Mol Sci. 24(1139)2023.PubMed/NCBI View Article : Google Scholar | |
|
Cheung EC and Vousden KH: The role of ROS in tumour development and progression. Nat Rev Cancer. 22:280–297. 2022.PubMed/NCBI View Article : Google Scholar | |
|
Zhang S, Wang C, Tang S, Deng S, Zhou Y, Dai C, Yang X and Xiao X: Inhibition of autophagy promotes caspase-mediated apoptosis by tunicamycin in HepG2 cells. Toxicol Mech Methods. 24:654–665. 2014.PubMed/NCBI View Article : Google Scholar | |
|
Yao N, Li YJ, Lei YH, Hu N, Chen WM, Yao Z, Yu M, Liu JS, Ye WC and Zhang DM: A piperazidine derivative of 23-hydroxy betulinic acid induces a mitochondria-derived ROS burst to trigger apoptotic cell death in hepatocellular carcinoma cells. J Exp Clin Cancer Res. 35(192)2016.PubMed/NCBI View Article : Google Scholar | |
|
Liu M, Zhao X, Xiao L, Liu G, Liu H, Wang X, Feng X and Lin X: Cytotoxicity of the compounds isolated from Pulsatilla chinensis saponins and apoptosis induced by 23-hydroxybetulinic acid. Pharm Biol. 53:1–9. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Yu C, Chen F, Jiang J, Zhang H and Zhou M: Screening key genes and signaling pathways in colorectal cancer by integrated bioinformatics analysis. Mol Med Rep. 20:1259–1269. 2019.PubMed/NCBI View Article : Google Scholar | |
|
Zhang Z, Zhang X, Meng L, Gong M, Li J, Shi W, Qiu J, Yang Y, Zhao J, Suo Y, et al: Pioglitazone inhibits diabetes-induced atrial mitochondrial oxidative stress and improves mitochondrial biogenesis, dynamics, and function through the PPAR-γ/PGC-1α signaling pathway. Front Pharmacol. 12(658362)2021.PubMed/NCBI View Article : Google Scholar |